Date Published:
2005
Abstract:
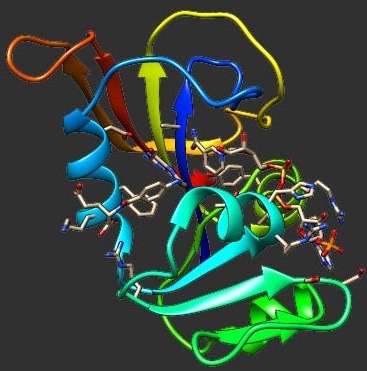
We use an integrated computational approach to reconstruct accurately the transition state ensemble (TSE) for folding of the src-SH3 protein domain. We first identify putative TSE conformations from free energy surfaces generated by importance sampling molecular dynamics for a fully atomic, solvated model of the src-SH3 protein domain. These putative TSE conformations are then subjected to a folding analysis using a coarse-grained representation of the protein and rapid discrete molecular dynamics simulations. Those conformations that fold to the native conformation with a probability (Pfold) of approximately 0.5, constitute the true transition state. Approximately 20% of the putative TSE structures were found to have a Pfold near 0.5, indicating that, although correct TSE conformations are populated at the free energy barrier, there is a critical need to refine this ensemble. Our simulations indicate that the true TSE conformations are compact, with a well-defined central β sheet, in good agreement with previous experimental and theoretical studies. A structured central β sheet was found to be present in a number of pre-TSE conformations, however, indicating that this element, although required in the transition state, does not define it uniquely. An additional tight cluster of contacts between highly conserved residues belonging to the diverging turn and second β-sheet of the protein emerged as being critical elements of the folding nucleus. A number of commonly used order parameters to identify the transition state for folding were investigated, with the number of native Cβ contacts displaying the most satisfactory correlation with Pfold values.
Website