Abstract:
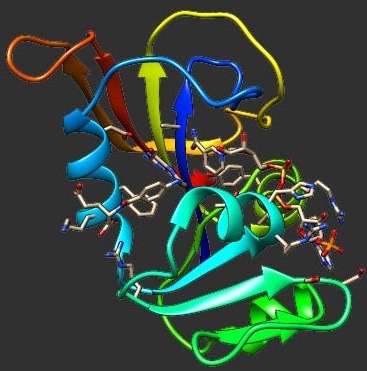
Directed evolution using random mutation in vast sequence space leads to the low probability of obtaining target proteins. Emerging engineering strategies with computational tools are developed for more trustable outcomes. We used some semi-rational design methods to modify an industrial enzyme, namely cellobiose 2-epimerase (CE). A mutant was selected for its better thermostability and isomerization activity. The tradeoffs between thermostability, epimerization activity and isomerization activity of the CE mutants were different. To investigate the computational prediction performance of protein stability upon point mutations, molecular dynamics (MD) simulation analyses were conducted. The root mean square deviation (RMSD) and hydrogen bond analyses reproduced the correct trends in stability changes of the wild-type and mutated CEs with relatively high accuracy (correlation coefficients r ~ 0.5–0.8). The simulation temperature and time are important factors that influence the prediction performance. Our result shows that thermostability predictors calculated from MD simulation do better in predicting the thermostability changes of the mutated enzymes than the predictors using static-state information of the enzymes.
Publisher's Version